1 Introduction
Fig. 1 (Color online) Schematic depiction of force sensor, fork generation, and the transition between fork state and kink state. (a) Force exerted on dsDNA and its components of shear force and compressive force. (b) Fork-structure generation unzipped by shearing force at the junction of dsDNA and ssDNA; $\Delta x$ is length of unzipped dsDNA corresponding to every fork generation. (c) Force sensor in fork state and kink state, the transition between the force sensor in fork state and the one in kink state. |
2 Model and Simulation Method
2.1 Force-Sensor Model
2.2 Simulation Method
3 Results
3.1 Generation of Forks at Junctions in Force Sensor
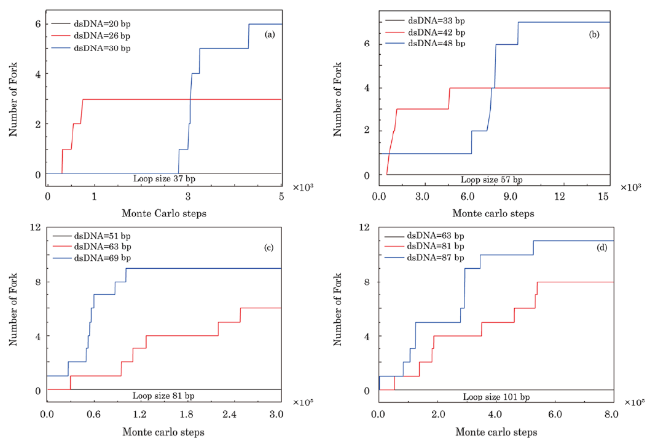
Fig. 2 (Color online) Fork generation at junction of dsDNA and ssDNA under conditions without kink generation in dsDNA. Loop lengths of the sensor in Figs. 2(a)-2(d) are 37 bp, 57 bp, 81 bp, and 101 bp, respectively, in which the loop is complemented by ssDNA with different lengths to form dsDNA; generation frequency of forks pertains to contour lengths of dsDNA in sensor proportionally. A critical contour length of dsDNA exists in every sensor and their values are 20 bp, 33 bp, 51 bp, and 63 bp, respectively. Only when the contour length of dsDNA in sensor becomes larger than the critical length does the fork structure begins to be generated. |
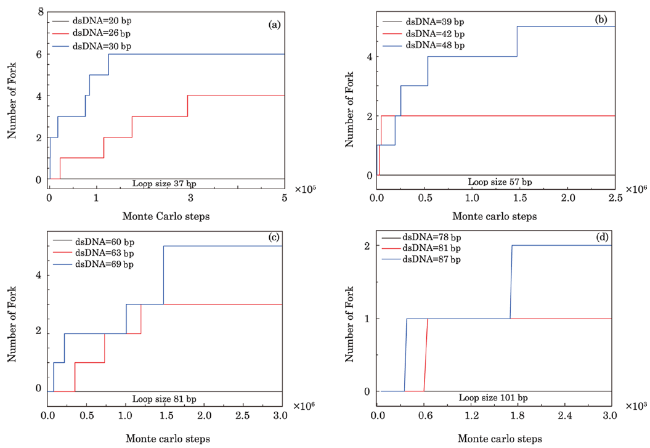
Fig. 3 (Color online) Fork generation at the junction of dsDNA and ssDNA under conditions with kink structure in dsDNA. Loop length of sensor in Figs. 3(a)-3(d) is 37 bp, 57 bp, 81 bp, and 101 bp, respectively, in which the loop is complemented by ssDNA with different lengths to form dsDNA; generation frequency of the fork pertains to contour length of dsDNA in the sensor proportionally. A critical contour length of dsDNA exists in every sensor and their values are approximately 20 bp, 39 bp, 60 bp, and 78 bp, respectively. Fork structure begins to be generated when the contour length of dsDNA in sensor becomes larger than the critical length. |
3.2 Forces Measured in Force Sensor
3.3 Effects of NN Stacking Interaction $\Delta\xi$ on Force Detection
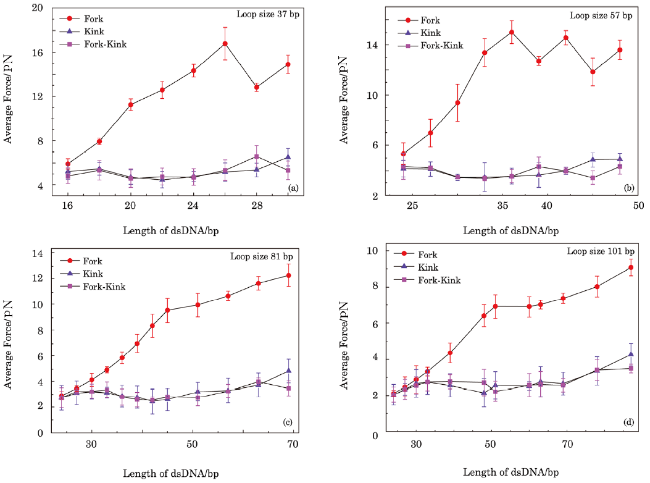
Fig. 4 Forces extracted from simulations as function of contour length of dsDNA in sensor. Three conditions are considered in the simulations, namely, that without kink structure in dsDNA but with a fork at the junction of dsDNA and ssDNA, that with kink structure in dsDNA but without a fork at the junction of dsDNA and ssDNA, and that with kink structure in dsDNA and a fork at the junction of dsDNA and ssDNA. NN base pair stacking interaction $\Delta\xi$ and excited energy $\mu$ used in simulations are $1.3k_{B}T$ and $8k_{B}T$, respectively. |
Fig. 5 Forces extracted from simulations as function of contour length of dsDNA in sensor. Three conditions are considered in simulations, namely that without kink structure in dsDNA but with a fork at the junction of dsDNA and ssDNA, that with kink structure in dsDNA but without a fork at the junction of dsDNA and ssDNA, and that with kink structure in dsDNA and a fork at the junction of dsDNA and ssDNA. NN stacking interaction $\Delta\xi$ and the excited energy $\mu$ used in simulations are $2.7k_{B}T$ and $8k_{B}T$, respectively. |
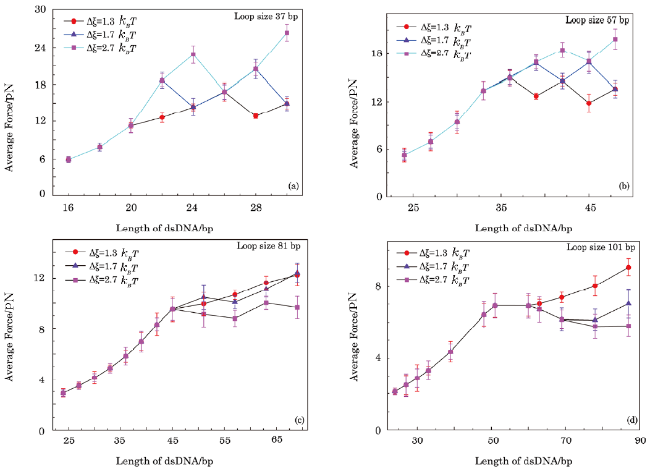
Fig. 6 Forces extracted from simulations under condition III as function of contour length of dsDNA in sensor, namely, that without kink structure in dsDNA but with a fork at the junctions of dsDNA and ssDNA. NN stacking interaction $\Delta\xi$ ranges from $1.3k_{B}T$ to $2.7k_{B}T$ and the excited energy $\mu$ used in simulations is $8k_{B}T$. |
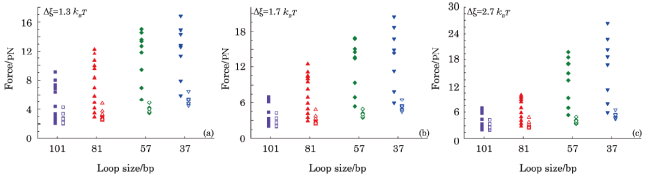
Fig. 7 All of the possible forces extracted from loops annealed by different-length ssDNA. NN stacking interaction $\Delta\xi$ for every figure are $1.3k_{B}T$, $1.7k_{B}T$, and $2.7k_{B}T$. Solid and empty symbols indicate the fork state and kink state, respectively. Each point corresponds to a distinct construct. |
3.4 State Transition between Fork and Kink
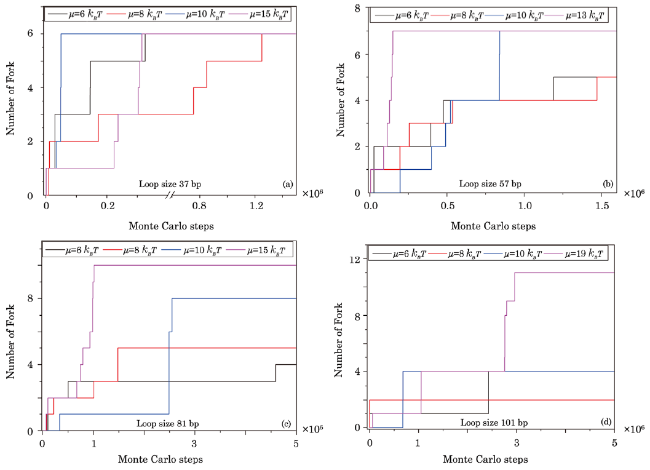
Fig. 8 (Color online) Effects of excited energy on fork structure generation. Loop lengths of sensor in Figs. 8(a)-8(d) are 37 bp, 57 bp, 81 bp, and 101 bp and complemented by 30 bp, 48 bp, 69 bp, and 87 bp ssDNA, respectively. Simulations are carried out under the condition with kink structure in dsDNA, fork at the junction between ssDNA and dsDNA, and NN stacking energy $\Delta\xi=1.3k_{B}T$. The fork in every sensor is generated more frequently with increasing excited energy. |
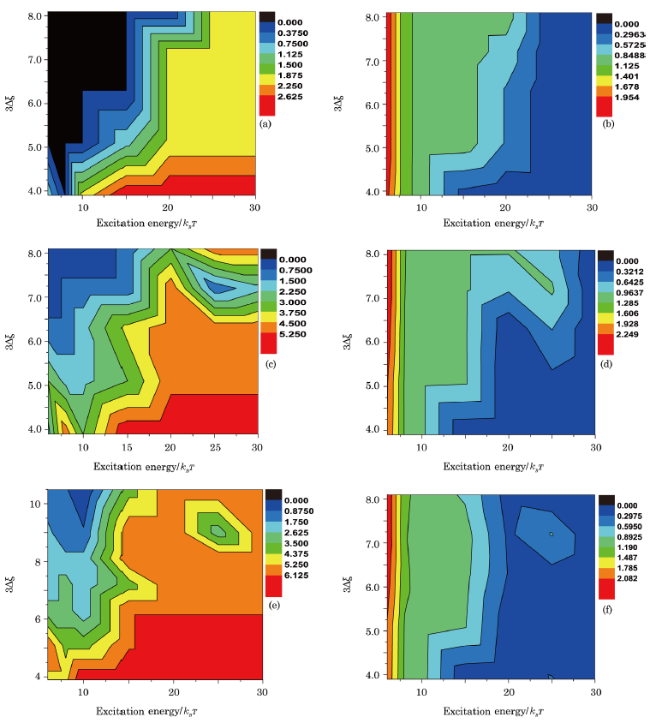
Fig. 9 (Color online) Phase diagram of fork ((a), (c), and (e)) and kink ((b), (d), and (f)) in the phase space of NN stacking energy $\Delta\xi$ and excited energy $\mu$. Color bar indicates the number of forks or kinks generated in the sensor with loop size 57 bp. From top to bottom, the sensor is complemented by ssDNA with lengths 39 bp, 45 bp, and 48 bp, respectively. |