1 Introduction
Fig. 1 (Color online) Structural demonstration of melittin and three representative gain-of-function variants, MelP1, MelP3 and MelP5. (a) Amino acid sequences of the peptides. Different types of residues are colored differently: grey for hydrophobic residues, orange for hydrophilic ones, while blue for charged ones. Residues in red are the mutated ones from parent melittin. Arrow "$N$" or "$C$" indicates the $N$ or $C$ terminus of the peptide. (b) Conformational structure of melittin and MelP5 (when binding on a membrane). The two $\alpha$-helixes of melittin are sketched with transparent cylinders, being connected with the kink part between them. Corresponding amino acid residues are highlighted with the same color scheme as in (a). |
2 Methods
2.1 Coarse-Grained Models
2.2 Coarse-Grained Simulations
2.3 Collective Variables (CVs)
2.4 MetaD Simulations
2.5 Minimum Free Energy Paths (MFEPs)
3 Results and Discussion
3.1 Peptide-Induced Membrane Disturbance and Poration
Fig. 2 (Color online) Membrane poration of different peptides at varying $P/L$ ratios. (a) Probability of membrane poration under the action of melittin and its variants with various $P/L$ ratios. For each parameter, three independent runs are performed for statistical analysis. Different colors are used to represent different types of pores. (b) Snapshots of two types of membrane pores. For clarification, only lipid head groups (blue: PO$_4$ group; orange: NC$_3$ group) and peptides are shown. (c) Time-dependent evolution of pore size under the action of melittin or MelP5, indicating the formation of transient or stable pore, respectively. Central lines are bold to guide eyes. (d), (e) Thinning and stretching (indicated by the area per lipid, APL) of lipid bilayer membranes under the action of peptides with various $P/L$ ratios. |
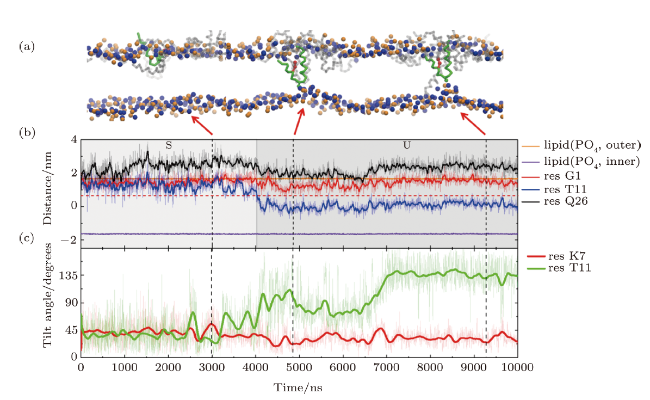
Fig. 3 (Color online) Formation process of a T-pore under the action of MelP5 at $P/L$=1/43. (a) Snapshots showing the interaction states, "S", "U" and "T", during the MelP5-membrane interaction process. The peptide which finally penetrate the membrane is tracked and colored red (with the kink residue T11 marked green), while the others are grey. (b), (c) Time-dependent evolution of the z-position (b) or tilt angle (c) of key residues, including G1 and Q26 at the two termini, T11 in the kink region, and the only charged residue K7 in helix-1. The z-position of lipid heads (PO$_4$ groups) of the two leaflets are shown as references of the membrane surfaces (solid orange and purple lines). |
3.2 Kinetics of Pore Formation
Fig. 4 (Color online) Residue-specialized interaction energy distribution between lipid bilayer and peptide in various states (i.e., "S", "U" or "T"). Left: melittin; Right: MelP5. In all cases, $P/L$=1/43. |
3.3 Free Energy Landscapes of the Peptide-Membrane Interactions
Fig. 5 (Color online) Different ability of different peptides, including melittin and its variants, to form "U" conformation during the membrane interactions. (a) Percentage of the peptides with U-shaped conformation during the membrane interaction process. Here, $P/L$=1/43. (b) Free energy profiles indicating the possibility to realize "U" conformation of each type of peptides. Inset shows definition of the CVs of $Z_N$ and $Z_{\rm kink}$. |
Fig. 6 (Color online) Free energy landscape of the transmembrane insertion of a single melittin or MelP5. (a) Landscape with the CVs of $Z_N$ and $Z_{\rm kink}$. (b) Corresponding minimum free energy pathways obtained from (a). Red lines demonstrate two leaflets of the membrane. $N$ terminus and residue 11 (at the kink region) are colored in red and green, respectively. |
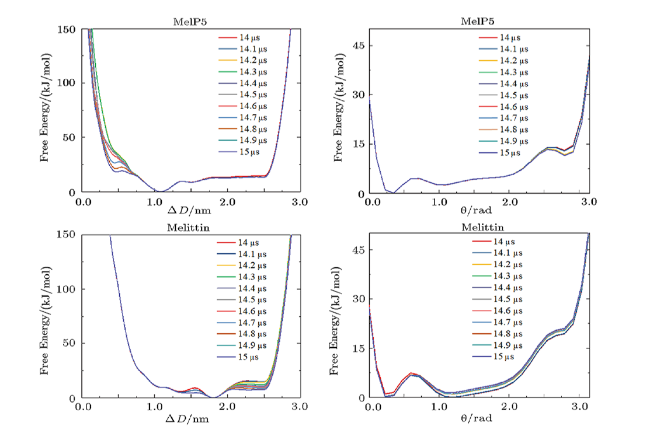
Fig. 7 (Color online) Aggregation of peptides on/in the membrane during the peptide-membrane interactions. (a) Evolution of peptide aggregation during the interaction process demonstrated by the cluster number (top) and the peptide number in the largest cluster (bottom). $P/L$=1/43. (b), (c) Free energy landscape of the aggregation of two peptides (b) and the projection along different CVs (c). Inset in (c) represents definition of the two CVs, $\Delta D$ and $\theta$, which are used to describe the aggregation condition of the two peptides. |
Fig. 8 (Color online) Free energy landscape of the interaction between membrane and two melittin peptides. (a) Landscape with the CVs of $Z_{\rm kink}$ values of the two peptides. (b) Energy projection along the CV of $Z_{\rm kink1}$. (c) Snapshots corresponding to the energy minima states in (a). |
4 Conclusion
Appendix
Table S1 Decrease of membrane thicknes(-∆h/h)×100% |
 |
Table S2 Increase of membrane area(-∆A/A)×100% |
 |
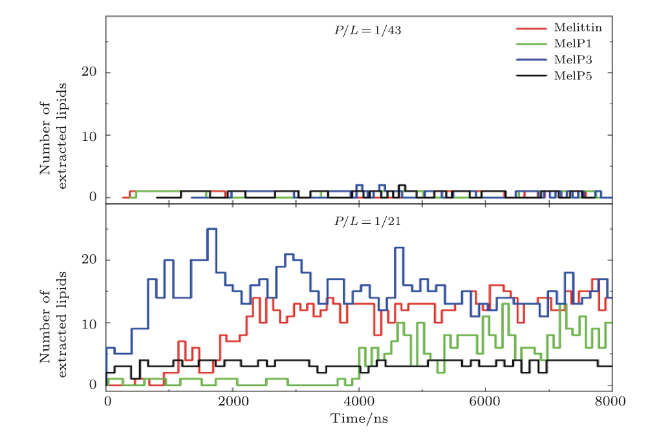
Fig. S1 (Color online) Number of lipids extracted from the bilayer membranes by melittin or its variants at $P/L$=1/43 (top) or 1/21 (bottom). Lipid extraction behavior occurs mostly at the high $P/L$ ratio. |
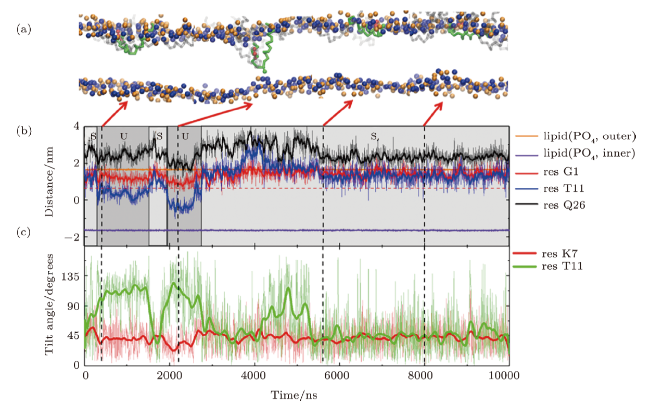
Fig. S2 (Color online) U-shaped MelP5 in a T-pore (the same trajectory as shown in Fig. 3). (a)Snapshots showing different interaction states between MelP5 and membrane (S and U). The peptide,which can have a U-shaped conformation, is colored with green, and the others are grey (includethe T-shaped MelP5). (b) Evolution of locations of termini (residues G1 and Q26) and kink part(residue T11) of the peptide (the green one in (a)). For comparison, the locations of lipid heads(PO4 groups) of outer and inner leaflet are also shown, respectively. (c) Changes of tilt angles (inset)of residues K7 and T11. For the U-shaped MelP5, flip of the side chain of residue T11 occurs while the configuration change of basic residue K7 does not, compared with the T-shaped MelP5 (see Fig. 3 in main text). The similarity in configuration changes of residue T11 and the difference in K7 indicate their roles in the formation of U-shape conformation of a peptide and the translocation of its $N$ terminus, respectively. $P/L$=1/43. |
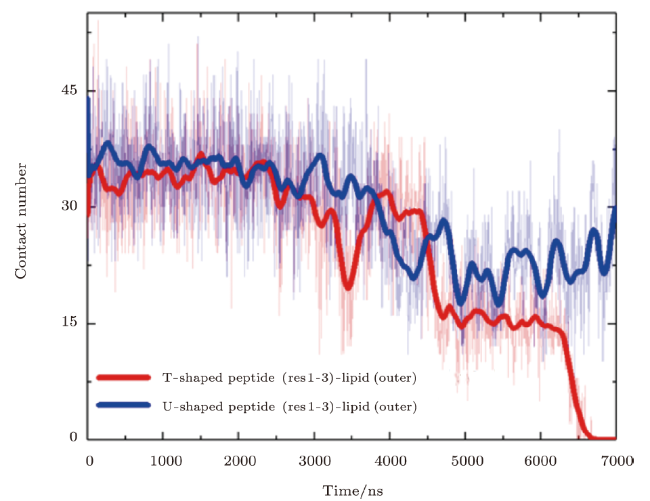
Fig. S3 (Color online) Contact situations of the $N$ terminus of a MelP5 with the transmembrane conformation (denoted as T-shaped) or a MelP5 with U-shaped conformation with the lipid heads in the outer leaflet of the bilayer. After the formation of U-shaped conformation ($\geq$4000 ns), the contact number between the $N$ terminus of the T-shaped MelP5 and lipid heads in the outer leaflet becomes obviously smaller than that of the U-shaped MelP5, and decrease to zero finally. This difference possibly caused by the peptide aggregation, along with flip of side chain of K7 of the peptide (as shown in Figs. 3 and S2), weakens the interaction between the $N$ terminus of the peptide and lipid heads, and facilitates the membrane translocation of the $N$ terminus of the peptide. $P/L$=1/43. |
Fig. S4 (Color online) Formation process of an unstable U-pore under the action of melittin at $P/L$=1/43. (a) Snapshots showing different interaction states between melittin and membrane ("S" and "U"). One U-shaped peptide is tracked and colored green (with residue K7 in red), while the others are grey. (b), (c) Time-dependent evolution of the z-position (b) and tilt angle (c) of key residues during the interaction process, including G1, Q26, T11, and K7. |
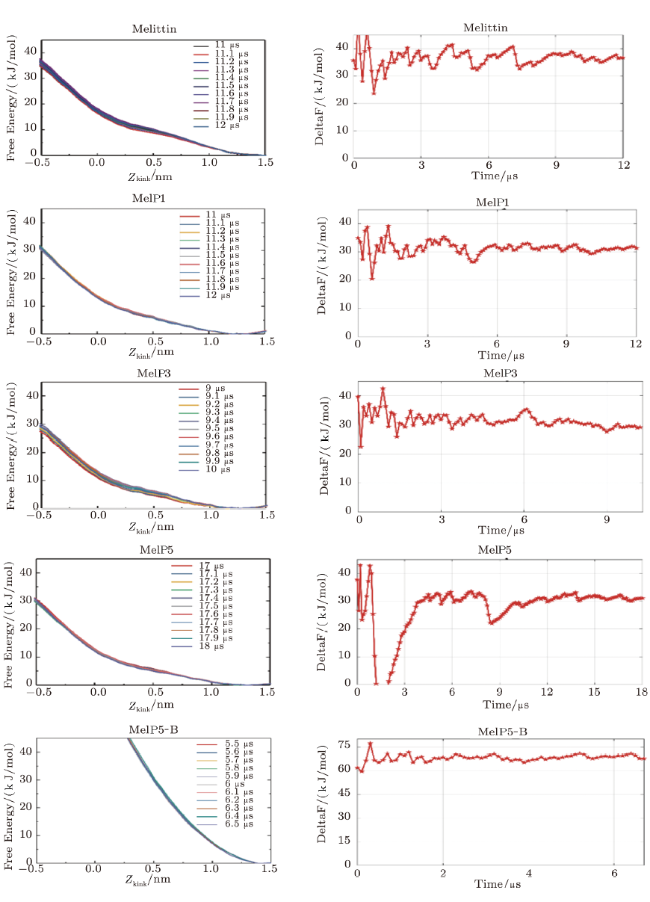
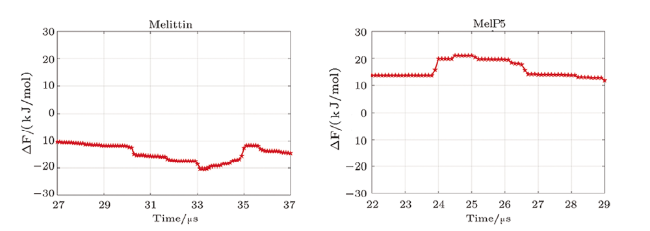
Fig. S5 (Color online) Convergence of the free energy of the formation of U-shaped peptides. Left: Evolution of the free energy profile with time; Right: the line refers to the free energy difference between the membrane binding state ($Z_{\rm kink}=1.2\sim1.5)$ and the U-shaped conformation state ($Z_{\rm kink}=-0.5$) of the peptide. For MelP5\_B. the convergence is judged by the free energy difference between the membrane binding state ($Z_{\rm kink}=1.2\sim1.5$) and that at $Z_{\rm kink}$=0. |
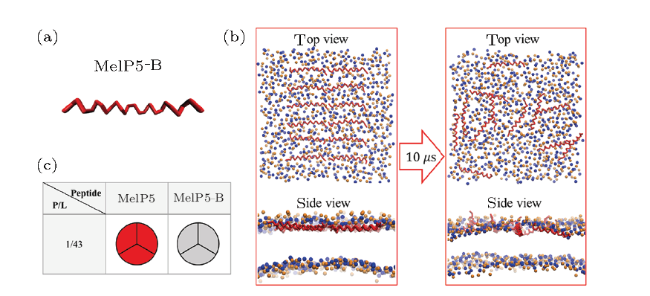
Fig. S6 (Color online) (a) Conformational structure of MelP5-B which is the structure-variant of MelP5. (b) Interaction states between MelP5-B and the lipid bilayer. (c) Formation probability of membrane pore under the action of MelP5-B at $P/L$=1/43. For each parameter, three independent runs are performed to make statistics. Red: T-pore; grey: no pore. Herein, control simulations are carried out to confirm the important role of the kink structure of the peptide. A variant of MelP5, named MelP5-B, is built. MelP5-B has the same amino acid sequence as MelP5 while with a full $\alpha$-helical chain structure, but without the non $\alpha$-helical kink region (Fig. S6(a)). Energy calculations show that it is rather difficult for this peptide to form a "U" conformation (dashed line in Fig. 6(b) in main text); simultaneously, transmembrane pore is hard to form (Figs. S6(b)-S6(c)). |
Fig. S7 (Color online) Convergence of the free energy of the transmembrane penetration of a melittin and MelP5.The lines refer to the free energy difference between the two saddle-points of two pathways (path U or T) in each free energy surface. The ranges of two saddle-points of two pathways are: for melittin, ($Z_{\rm kink}=(1.21\sim1.27$) nm, $Z_N=(0\sim0.15)$ nm) and ($Z_{\rm kink}=(-0.92\sim-0.87$) nm, $Z_N=(-0.4\sim-0.2$) nm); for MelP5, ($Z_{\rm kink}=(1.17\sim1.21$) nm, $Z_N=(-0.1\sim0.03$) nm) and ($Z_{\rm kink}=(-0.8\sim-0.75$) nm, $Z_N=(0.15\sim0.25)$ nm). The fluctuation range of $\Delta F$ is less than 10 KJ/mol. |
Fig. S8 (Color online) Convergence of the free energy of the aggregation of two peptides on a membrane surface. Lines show the evolution of the free energy profile along with different CVs. |
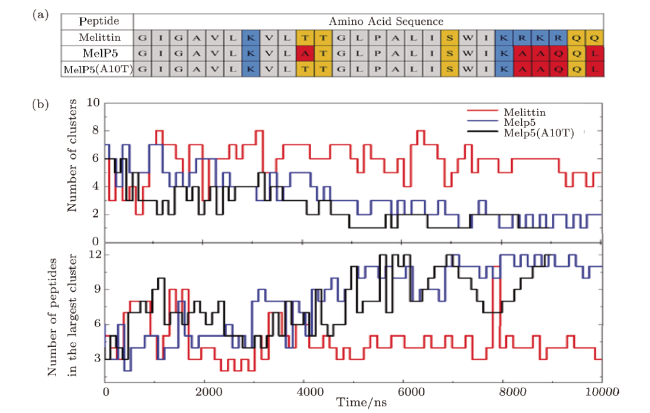
Fig. S9 (Color online) (a) Amino acid sequences of a variant of MelP5, named MelP5'(A10T).Color codes are same as those in Fig. 1 in the main text. (b) Peptide aggregation on the membrane surface indicated by cluster number (top) and the peptide number in the largest cluster (bottom).$P/L=43$. The results shown here show the aggregation situation of MelP5'(A10T). It is found that,although the 10th residue is changed back from A to T in MelP5'(A10T) (Fig. S9(a)),its aggregation situation is almost the same as that of MelP5 (Fig. S9(b)). |
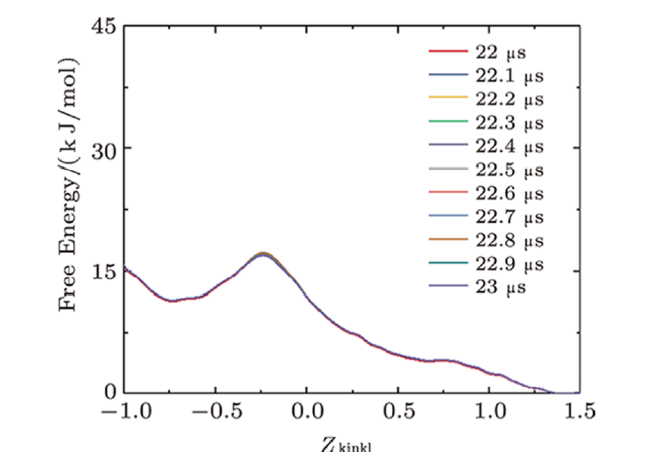
Fig. S10 (Color online) Convergence of the free energy of the interaction between two melittins and the membrane. Lines show the evolution of the free energy profile along with CV of $Z_{\rm kink1}$. |