Molecular dynamics (MD) simulation of biological macromolecules, beginning in 1977, have opened a new era "from structure to function" for the research of biophysics.
[1] MD simulation can provide the kinetic and thermodynamics information of proteins at the atomic level and reveal the important roles that protein dynamics played in many physiological processes.
[2]-[8] In addition, MD also makes up the shortcoming of the experiments and has becoming a significant complementary tool in explaining the experimental phenomenon and uncovering the experimental nature. Predicting and guiding the process of experiment in theory, importantly, MD simulation has been successfully applied to many fields due to its rapid development.
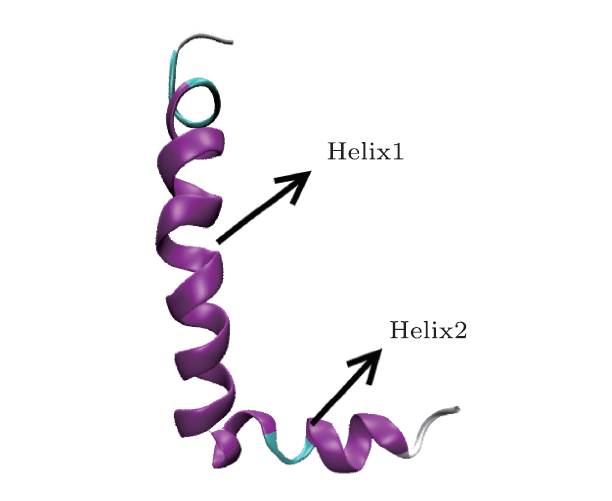
[9]-[13] Among them, the milestone event is the study for protein folding. Duan and Kollman firstly performed a 1 $\mu$s folding simulation for Villin headpiece with 36 residues in explicit solvent using AMBER96 force field, and they obtained the middle state, which was similar with the native state.
[14] The possible folding pathway of this protein is proposed in their study and from then that set off a folding simulation boom with MD. Shaw group has developed a specialized supercomputer to simulate the folding of some protein, which provides a new platform for solving a series of problems about protein folding using MD simulation technology.
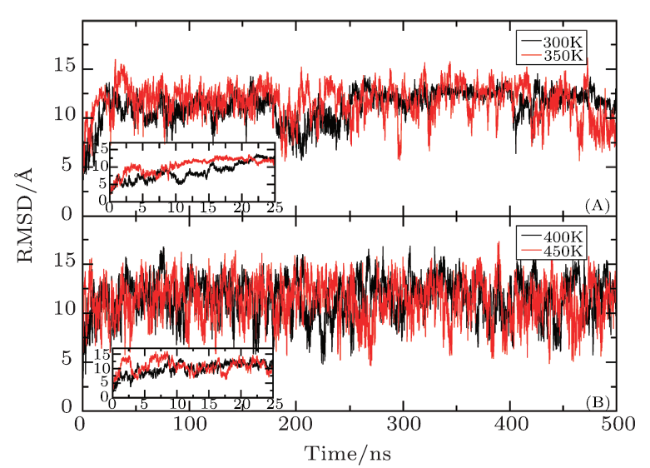
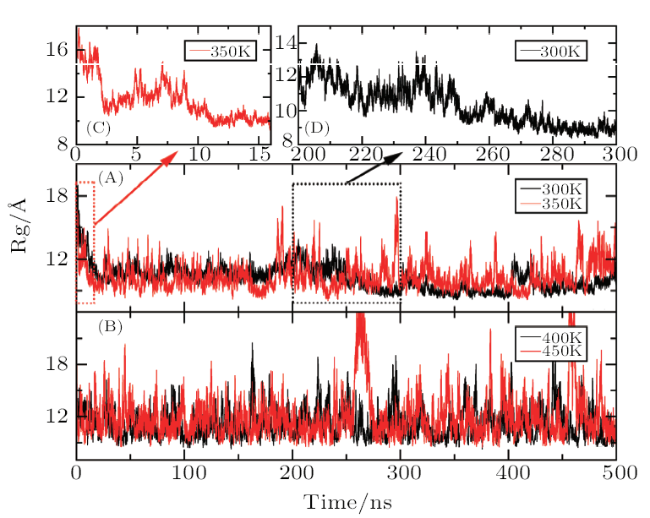
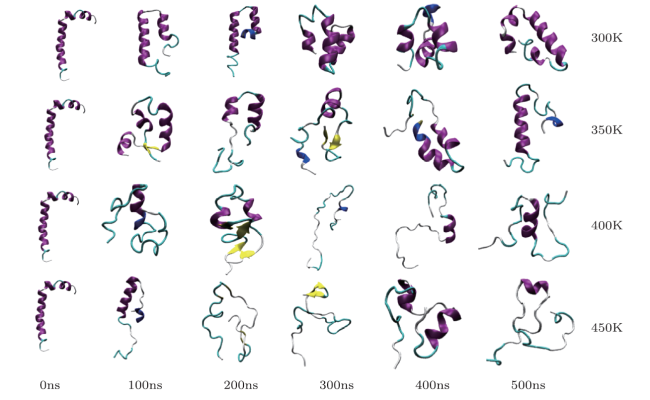
[15]-[16] However, due to the limited computational power of computer, MD simulation is often hindered by poor phase-space sampling efficiency on bumpy potential energy surfaces. It is difficult to traverse the thorough sampling of the entire phase space in the limited simulation time, which results into that the large-scale conformational changes usually are not observed.