1. Introduction
2. Methods
2.1. All-atom MD simulations
Table 1. The DNA sequences used in the work. |
| Name | Sequencesa | Cell size (nm3)b | Tmc |
|---|---|---|---|
| AA-TT | 5′-CGAAAAAAAAAAAAAAAAGC-3′ | 12 × 12 × 10 | 87.2 °C |
| GCTTTTTTTTTTTTTTTTCG | |||
| AT-TA | 5′-CGATATATATATATATATGC-3′ | 12 × 12 × 10 | 79.9 °C |
| GCTATATATATATATATACG | |||
| GENE | 5′-CGACTCTACGGCATCTGCGC-3′ | 12 × 12 × 10 | 104.2 °C |
| GCTGAGATGCCGTAGACGCG |
aThe sequences were selected according to the existing experiments [41, 51] and simulations [42]. | |
bThe cell sizes were used in our molecular dynamic simulations. | |
cThe melting temperatures are calculated at 0.15 M NaCl through the nearest-neighbor model with the measured thermodynamic parameters and the salt extension from [53, 59, 73]. |
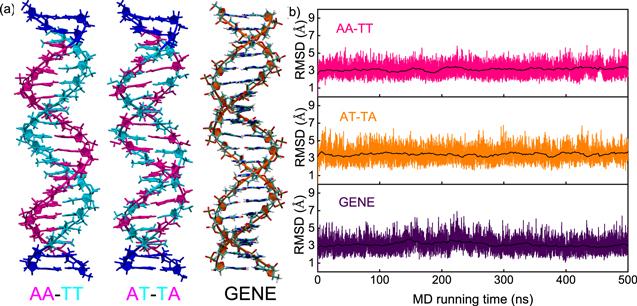
Figure 1. (a) The initial three-dimensional DNA structures with typical sequences of poly(A)-poly(T) duplex (AA-TT), poly(AT)-poly(TA) duplex (AT-TA) and generic one (GENE) for our all-atom molecular dynamics simulations, respectively. The three sequences were chosen according to the recent experiments [41, 51] and coarse-grained simulations [42]. (b) The root mean square deviations (RMSD) versus MD running time (500 ns) for the central 14-bp segment of the DNAs with three typical sequences. The black lines represent the RMSD values averaged over every 2 ns. The DNAs are in 150 mM NaCl solutions. |
2.2. Helical parameters and persistence length
2.3. Electrostatic calculations
2.4. A grooved coarse-grained model for DNA
3. Results and discussion
3.1. Persistence length of AA-TT is apparently larger than those of AT-TA and GENE sequences
Figure 2. (a) The bending angle distributions p(θ, l) versus bending angle θ over 10 base steps for the central 14-bp segments of the DNAs with three sequences of AA-TT, AT-TA and GENE. The bending persistence lengths can be estimated by fitting the shown curves to equation ( |
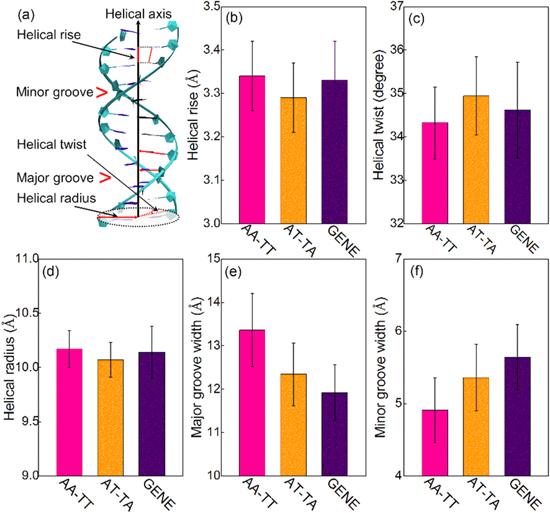
Table 2. The helical parameters and persistence length for DNAs with different sequencesa. |
| AA-TT | AT-TA | GENE | |
|---|---|---|---|
| Helical rise (Å) | 3.34 ± 0.08 | 3.29 ± 0.08 | 3.33 ± 0.09 |
| Helical twist (degree) | 34.32 ± 0.83 | 34.94 ± 0.90 | 34.62 ± 1.10 |
| Helical radius (Å) | 10.17 ± 0.17 | 10.07 ± 0.16 | 10.14 ± 0.24 |
| Major groove width (Å) | 13.36 ± 0.84 | 12.34 ± 0.72 | 11.92 ± 0.64 |
| Minor groove width (Å) | 4.91 ± 0.45 | 5.36 ± 0.46 | 5.64 ± 0.45 |
| Persistence length (nm)b | 63 | 48 | 49 |
3.2. Electrostatic energy makes the major contribution to sequence-dependent bending energy
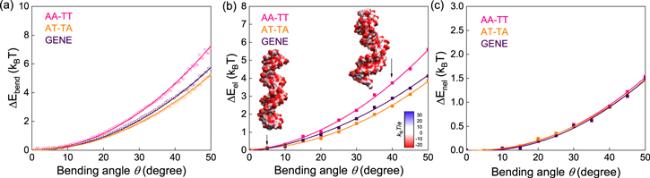
Figure 3. (a) The bending energy ΔEbend(θ) versus bending angle θ over the central 14 base pairs, for AA-TT, AT-TA and GENE. (b) The electrostatic bending energy ΔEel(θ) versus bending angle θ over the central 14 base pairs for AA-TT, AT-TA and GENE. Here, ΔEel(θ) was calculated directly with the APBS [68]. (c) The non-electrostatic bending energy ΔEnel(θ) versus bending angle θ over the central 14 base pairs for AA-TT, AT-TA and GENE. The electrostatic energies and the surface potentials in panels (b), (c) were calculated with the APBS [68]. |
3.3. Narrower minor groove width visibly increases electrostatic bending energy of AA-TT
Figure 4. (a) An illustration for helical rise, helical twist, helical radius, major groove width and minor groove width for a DNA. The helical parameters for the central 14-bp segments of AA-TT, AT-TA and GENE including helical rise (b), helical twist (c), helical radius (d), major groove width (e), minor groove width (f). The error bars are the standard deviations to the respective mean values. |
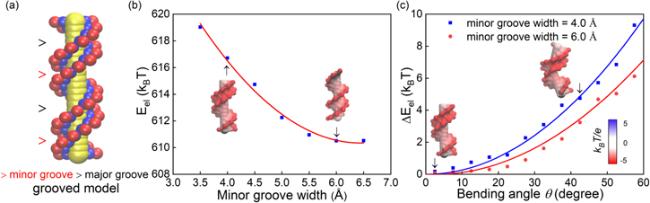
Figure 5. (a) A grooved coarse-grained model for DNA structure [71, 72], where every base pair is composed of two phosphate spheres (in red), two sugar spheres (in blue) and one big base sphere (in yellow); see details in the appendix. (b) The electrostatic energy for straight 14-bp DNAs (bending angle θ = 0) with different minor groove widths from the grooved DNA model. (c) The electrostatic bending energy ΔEel(θ) for a 14-bp DNA versus bending angle θ over central 6 base pairs. The electrostatic energies and the surface potentials in panels (b), (c) were calculated with the APBS [68]. |
Table 3. Effects of helical parameters on electrostatic energy for the grooved DNA modela. |
| Helical rise | Helical twist | Helical radius | Minor groove width | |
|---|---|---|---|---|
| 3.2 → 3.4 Å | 33.0 → 35.0° | 10.0 → 10.2Å | 4.0 → 6.0 Å | |
| Eel(θ = 0°) (kBT) | 614 ↓ | 610 ↓ | 611 ↓ | 617 ↓ |
| 609 | 609 | 610 | 610 | |
| △Eel(θ = 40°) (kBT) | 3.7 ↓ | 3.4 ↓ | 1.9 ↑ | 4.6 ↓ |
| 0.8 | 1.3 | 4.0 | 3.2 |
aThe bending angle θ is over 14-bp DNA, and the positive and negative effects by the changes of helical parameters are marked with ‘↑'and ‘↓', respectively. |