Understanding sequence effect in DNA bending elasticity by molecular dynamic simulations
|
Understanding sequence effect in DNA bending elasticity by molecular dynamic simulations |
| Xiao-Wei Qiang,Hai-Long Dong,Kai-Xin Xiong,Wenbing Zhang,Zhi-Jie Tan |
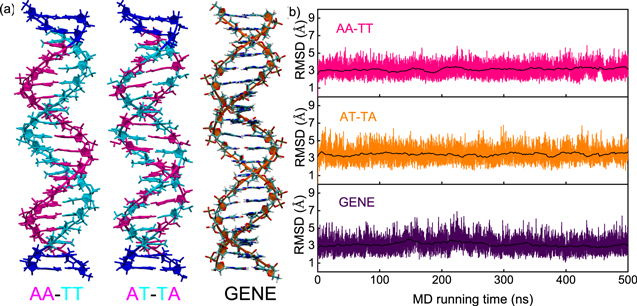
| Figure 1. (a) The initial three-dimensional DNA structures with typical sequences of poly(A)-poly(T) duplex (AA-TT), poly(AT)-poly(TA) duplex (AT-TA) and generic one (GENE) for our all-atom molecular dynamics simulations, respectively. The three sequences were chosen according to the recent experiments [ |

|