1. Introduction
2. Model
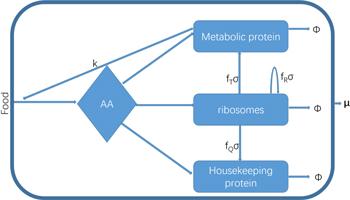
Figure 1. A coarse-grained model of the cell. Extracellular food is transported into the cell and is decomposed into amino acids (AA) by metabolic proteins. Amino acids are assembled into various proteins by ribosomes and are allocated to metabolic proteins, ribosomes, and housekeeping proteins reasonably to get a maximal growth rate. → φ means the degradation of proteins. μ is the growth rate. |
3. Deterministic results
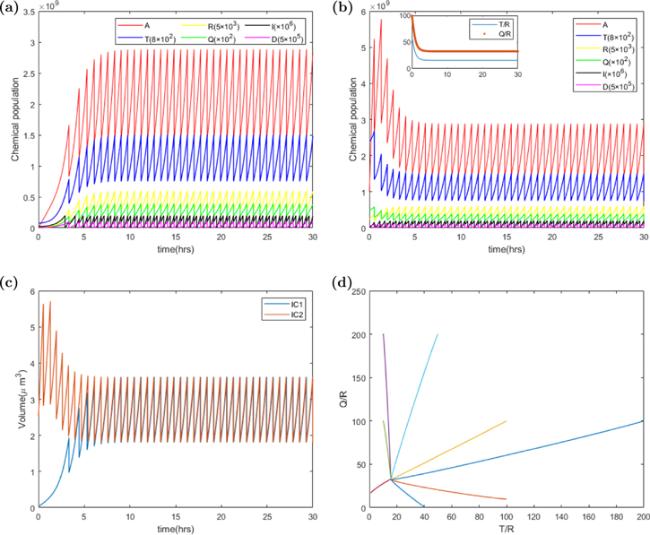
Figure 2. The deterministic trajectories of chemical populations, P, T, R, Q, I, D and volume V versus time t. Parameters of the ATRQID system are k = 3000 h−1, Σ = 3 ∗ 10−5 μm3 h−1, KD = 4 ∗ 10−12 h−1, KI = 6 ∗ 10−12 h−1, dR = dQ = dT = 0.1 h−1, Dc = 200, Ic = 200, Dr = 0, Ir = 0, and the other parameters of the model are shown in the table 1. The population values in (a) and (b) have been adjusted to make all of populations shown on the same figure by some multiplier factors. (a) The trajectory of populations versus time in a cell with the initial condition IC1: A(0) = 103, T(0) = 105, R(0) = 103, Q(0) = 104, I(0) = 20, D(0) = 10. The populations’ dynamics evolve to a steady state, and the chemical populations in the daughter cell at the birth are Ab = 1.28 ∗ 109, Tb = 8.39 ∗ 105, Rb = 5.26 ∗ 104, Qb = 1.71 ∗ 106, Ib = 88, Db = 0. The doubling time is 0.7235 h. (b) The trajectory of populations versus time in a cell with the initial condition IC2: A(0) = 109, T(0) = 3∗106, R(0) = 5∗104, Q(0) = 5∗106, I(0) = 10, D(0) = 100. The population dynamics reach a same steady state as IC1. (c) The plot of cell size versus time on the two initial conditions IC1 and IC2. (d) The 2D space of Q/R and T/R shows that the trajectories converge to the same attractor independent of initial conditions. |
Table 1. The dynamics parameters and biological constants in the ATRQID system. |
| Constants/Parameters | Symbol | Value | Reference |
|---|---|---|---|
| Protein density | g | 0.25 g cm−3 | [76] |
| Molecular weight per ribosome R | nR | 7336aa | [75] |
| Molecular weight per metabolic protein T on average | nT | 325aa | [70] |
| Molecular weight per housekeeping protein P on average | nQ | 325aa | [70] |
| Protein degradation rate respectively | dR, dT, dQ | Given | [71, 72] |
| The efficiency of metabolic enzymes T | k | Given | |
| Ribosomal catalytic efficiency | Σ | Given | |
| The fractions of ribosomes engaged in making ribosomes R | fR | Optimized by growth rate | Equation ( |
| The fractions of ribosomes engaged in making metabolic protein T | fT | Optimized by growth rate | Equation ( |
| The fractions of ribosomes engaged in making housekeeping protein T | fQ | 0.45 | [73] |
| DNA replication initiation protein per origin threshold | Ic | Given | |
| Cell division initiation protein threshold | Dc | Given | |
| The average mass of amino acids | mA | 3000Da | [74] |
| DNA replication initiation protein synthesis rate constant | KI | Given | |
| Cell division initiation protein synthesis rate constant | KD | Given |
4. Stochastic numerical results
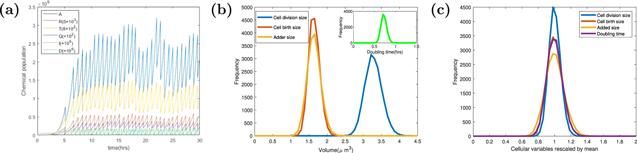
Figure 3. The stochastic trajectories and the scale-free property of cell size. (a) The stochastic trajectories of chemical populations, P, T, R, Q, I, D and volume V versus time t. Parameters of the dynamics systems are k = 3000 h−1, Σ = 3 ∗ 10−5 μm3 h−1, KD = 4 ∗ 10−12 h−1, KI = 6 ∗ 10−12 h−1, dR = dQ = dT = 0.1 h−1, Dc = 200, Ic = 200, Dr = 0, Ir = 0, and the other parameters of the model are shown in the table 1. The population values in figure 3(a) have been adjusted to make all of populations shown on the same figure by some multiplier factors. (b) The frequency distribution of cell division size, birth size, added size, and inter-division time in the steady state. The average division cell size ⟨Vd⟩ is 3.279 μm3, average birth cell size ⟨Vb⟩ = 1.642 μm3, average added cell size ⟨Δ⟩ = 1.637 μm3, average division time ⟨τ⟩ = 0.723 h. (c) The frequency distribution of cellular variables is re-scaled by their respective means. The curves denoted by cell division size, cell birth size, added size and doubling time stand for Vd/⟨Vd⟩, Vb/⟨Vb⟩, Δ/⟨Δ⟩ and τ/⟨τ⟩. The coefficient of variation of those cellular variables are 0.071, 0.093, 0.113, 0.094. |
Figure 4. The cell birth size distribution versus cell size distribution of lineage data. (a) In the same lineage, the distribution of tens of thousands of consecutive cells at birth. Three distributions are used to fit this distribution, namely: Gaussian distribution, gamma distribution, and lognormal distribution. (b) In the same lineage, the cell size distribution of cell lineage data for dozens of consecutive generations. |
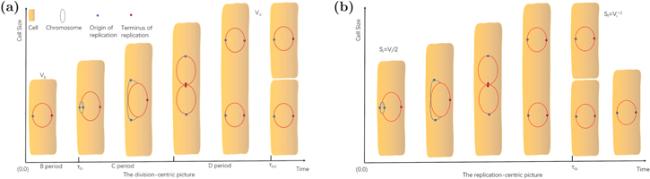
Figure 5. The division-centric picture and the replication-centric picture of the cell cycle. (a) In the division-centric picture, the cell cycle is separated into three periods. The first period is from cell birth to initiation of DNA replication, named B period. The second period is the time interval DNA replication occupies, called C period. The last one is from the completion of the chromosome of replication to cell division, named D period. DNA replication begins at the origin when the cell size reaches a critical volume Vi and ends at the terminus. (b) The replication-centric picture is defined as two consecutive chromosome replication initiations. It is worth noting that (b) is only tenable at a slow growth rate. When doubling time is shorter than C + D, there will be more than one round replication in the cell. All the biological variables are defined in table 2. |
4.1. The adder property of division-centric picture
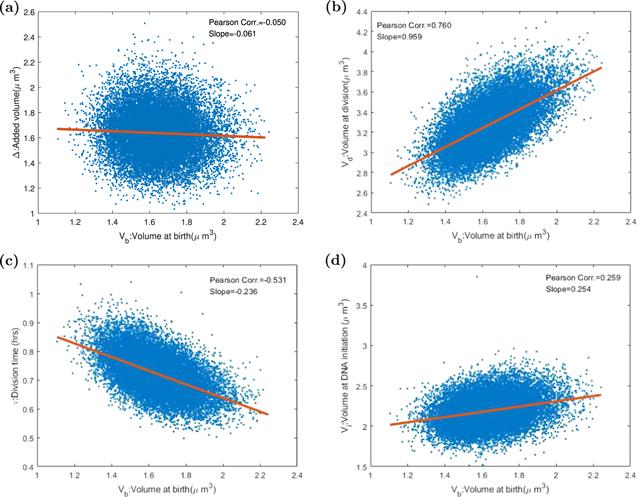
Figure 6. The Pearson correlation coefficients and slopes between various variables in the division-centric picture. Each point stands for one cell in 20 000 consecutive divisions after the system gets into the steady state. The red line is the linear fitting curve. (a) The X-axis refers to the cell birth volume, and the Y-axis refers to the increased volume during the division cycle. In the ideal adder model, the correlation coefficient of Vb and Δ should be 0. Our result is −0.050, so that add property is proved in our model. (b) The correlation coefficient of Vb, volume at birth time, and Vd, volume at division time, is 0.760. The slope of the fitting curve is about 1. (c) The doubling time τ is negatively correlated with birth size. In the ideal adder model, the correlation coefficient should be −0.5, compared with our model −0.531. (d) The replication initiation size Vi is positively correlated with Vb with the correlation coefficient being 0.259. If Donchie’s hypothesis is correct, the value should be 0. |
4.2. The adder property of replication-centric picture
Table 2. Variables definitions. |
| Variables | Symbol |
|---|---|
| Size at birth | Vb |
| Size at division | Vd |
| Doubling time | τ |
| Size at replication | Vi |
| initiation | |
| Duration between birth | τbi |
| and replication | |
| Size per origin at initial | Si |
| replication initiation | |
| Size per origin at final | Sf |
| replication initiation | |
| Duration between consecutive | τif |
| replication initiations | |
| Size per origin at birth | Sb |
| Duration between replication | τib |
| initiations and birth | |
| Cell growth rate | $\mu =1/\tau {\rm{ln}}({V}_{d}/{V}_{b})$ |
| Division added size | Δ = Vd − Vb |
| Replication added size | Δif = Sf − Si |
| Birth-to-initiation added size | Λbi = Vi − Vb |
| Initiation-to-birth added size | Λib = Sb − Si |
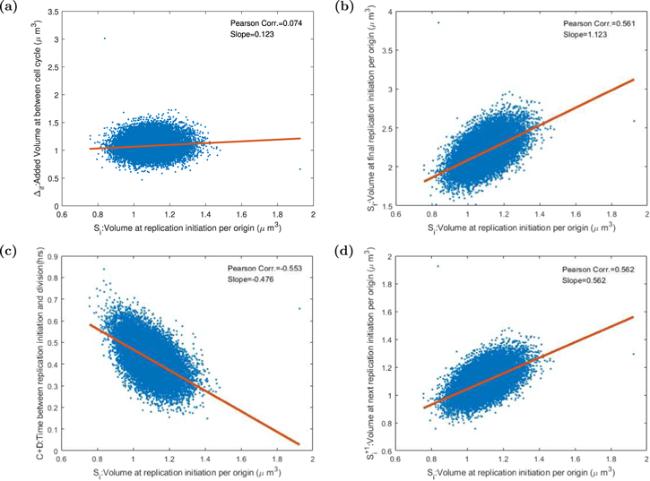
Figure 7. The Pearson correlation coefficients and slopes between various variables in the replication-centric picture. Each point stands for one cell in 20 000 consecutive cells after the system gets into the steady state. The red line is the linear fitting curve. (a) The increment between cell replication cycle Δif is uncorrelated with volume Si at replication initiation per origin. This result certificates the adder property in the replication-centric picture. (b) The correlation coefficient between the volume Si at the initial replication initiation per origin and the volume Sf at the final replication initiation per origin is 0.561 and the slope of the curve 1.123. (c) The time between replication initiation and cell division C + D is negatively correlated with Si. (d) The auto-correlation coefficient of Si is 0.562. If Donchie’s hypothesis is right, the value should be 0. |
4.3. Oscillation of DNA initiation protein expression
Figure 8. The Pearson correlation coefficients and slopes between birth size, replication initiation size, and added size in the replication-centric picture and division-centric picture when DNA initiation proteins are expressed periodically. Parameters of the dynamics systems are k = 3000 h−1, Σ = 3 ∗ 10−5 μm3 h−1, KD = 4 ∗ 10−12 h−1, KI = $9.2\ast {10}^{-12}\ast | \sin (t\ast \pi /1.446)| /\mathrm{hr}$, dR = dQ = dT = 0.1 h−1, and the other parameters of the model are shown in the table 1. (a) The mean added volume is unchangeable whatever the birth size is, in conflict with co-regulation theory. (b) The periodical oscillation expression of chromosome replication protein changes the initiation mass, destroying the replication adder. |
4.4. Oscillation of division protein expression
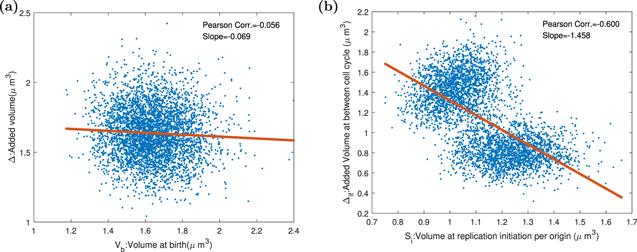
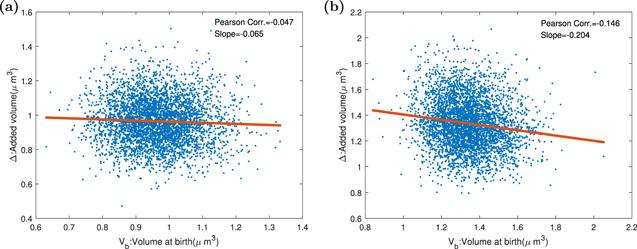
Figure 9. The Pearson correlation coefficients and slopes between birth size, replication initiation size, and added size in the replication-centric picture and division-centric picture when division proteins are expressed periodically. Parameters of the dynamics systems are k = 3000 h−1, Σ = 3 ∗ 10−5 μm3 h−1, ${K}_{D}=4\ast {10}^{-12}\ast | \sin (t\ast \pi /1.446)| /\mathrm{hr}$, KI = 3.4 ∗ 10−12 h−1, dR = dQ = dT = 0.1 h−1, and the other parameters of the model are shown in the table 1. (a) The oscillation of division protein destroys adder property. The smaller the birth size is, the bigger the adder size. (b) DNA replication initiation is not influenced by periodical division protein production, and replication adder is kept. |
4.5. Deviation from adder toward sizer at slow growth rate
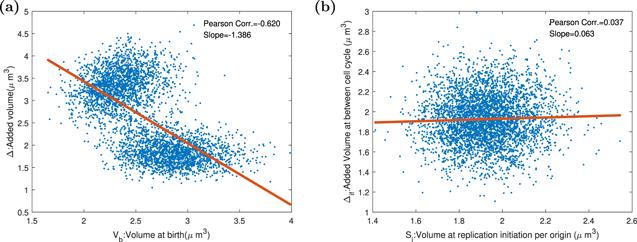
Figure 10. The Pearson correlation coefficients and the slopes between birth size and added size when division protein degradation rate is zero or not zero, and cells have an auto-inhibition mechanism. Parameters of the dynamics systems are k = 3000 h−1, Σ = 3 ∗ 10−5 μm3 h−1, KD = [200/(200 + d)] ∗ 10−11 h−1, KI = 7 ∗ 10−12 h−1, dR = dQ = dT = 0.1hr−1, and the other parameters of the model are shown in the table 1. (a) The degradation rate of division protein dD is 0. Cells represent adder property. (b) The degradation rate of division protein dD is 1hr−1. Cells slightly deviate from adder property to sizer property. |